| Page last updated
2 July 2019 |
MUSSELpdb: The MUSSEL Project Database
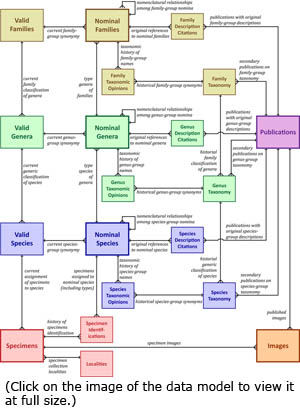 The peculiar history of freshwater mussel taxonomy — grossly over-named taxa at all levels, high (actual) global diversity, etc. — has made the management of the diversity data for the Unionoida cumbersome to say the least. The Entity Relationship Diagram (ERD) or schematic to the left depicts the data model that we have implemented to manage the various kinds of information that we need to track and verify. Click here to view a full-sized version of this diagram. The peculiar history of freshwater mussel taxonomy — grossly over-named taxa at all levels, high (actual) global diversity, etc. — has made the management of the diversity data for the Unionoida cumbersome to say the least. The Entity Relationship Diagram (ERD) or schematic to the left depicts the data model that we have implemented to manage the various kinds of information that we need to track and verify. Click here to view a full-sized version of this diagram.
For data entry and management, we have developed the MUSSELpdb in FileMaker Pro, beginning with version 5.5 and currently working in FileMaker Pro 17. The world wide web version of our database is powered by Google. In FileMaker, our relational database is composed of several files, most with multiple tables, and these are color-coded in the schematic of our data model (left): Families (yellow), Genera (green), Species (blue), Specimens (red), Publications (purple) and Images (orange). Functionally, the three taxonomic levels are built similarily. The other three files (Publications, Specimens, and Images) represent libraries of data utilized throughout the database, although Specimens and Images both connect directly to the taxon tables only at the Species-group level.
Serving to World Wide Web
One way to implement the MUSSELpdb would be solely via the World Wide Web. That plan would have certain advantages, the greatest of which would be a central, single copy of the database and live updates. However, there are also limiting drawbacks. For example, in the field (i.e., streamside) it often difficult for us to access the Internet. In addition, FileMaker Pro is powerful and worked very well for us and facilitated database development — there is no need to re-invent the wheel.
The legacy version of the MUSSELpdb was powered by MySQL (Structured Query Language) and PHP (PHP: Hypertext Preprocessor). Unfortunately, we have little institutional support for those two protocols. To facilitate HTML development and improve the frequency of data updates, the current version of the MUSSELpdb utilizes Python to create static web pages that are indexed and searchable with a custom Google search.
Taxonomic Tables
As each of the taxonomic levels are similarly structured, a discussion of the Species tables (blue) will be illustrative of all three. The Species tables all center around Nominal Species. That table contains original combinations for species-group level taxa, both available nomina and things like spelling errors, nude names, etc. The Valid Species table contains records for subsequent combinations of species we consider to be valid. Records in Nominal Species are linked to those in Valid Species, creating a one-to-many relationship between each valid species and the nomina synonymized under it. Each record in Valid Species is in turn linked to a valid genus above it. Thus, the Nominal Species table contains the objective entities that our classification of freshwater mussels is trying to arrange, and Valid Species implements that classification.
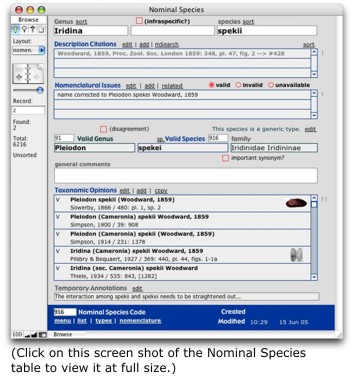 Associated with Nominal Species is further objective data. Species Descriptions contains citations to original descriptions and figures of each nomen. If type specimens are mentioned in the original description, these are also stored here. These data require a one-to-many relationship because there are often multiple, separate publications relevant to a species description. For example, Isaac Lea regularly first described a species only in a latin and without an illustration. It would be in a subsequent paper that the species would be figured and discussed. Species Nomenclature is the means by which records in Nominal Species can be linked to other records in the same table — e.g., if one nomen is a new name for another. Associated with Nominal Species is further objective data. Species Descriptions contains citations to original descriptions and figures of each nomen. If type specimens are mentioned in the original description, these are also stored here. These data require a one-to-many relationship because there are often multiple, separate publications relevant to a species description. For example, Isaac Lea regularly first described a species only in a latin and without an illustration. It would be in a subsequent paper that the species would be figured and discussed. Species Nomenclature is the means by which records in Nominal Species can be linked to other records in the same table — e.g., if one nomen is a new name for another.
We are not the first in history to try and revise the Unionoida. Not only have the methods and philosophies used to concoct arrangements changed over time, but various authors have disagreed as to how the various nomina should be arranged — which nomina are valid, in which genera they should be placed and which species are synonyms of what. Thus, the taxonomic histories of each name are useful for understanding how they have been treated in the past. Once the past has been captured and made explicit, we can move forward!
Each record in Species Taxonomies lists a subsequent combination of a particular author. This table is very like our Valid Species table except that it records how species were treated by others. Each record in Species Taxonomies is linked to one or more records in Species Synonymies, which serves as an associative entity between Nominal Species and Species Taxonomies. It is also in the Taxonomies tables that we store published information about types from the various authors. Of course, types are suppose to be objective entities representing particular nomina, but authorities frequently disagree about what entity is actually the type!
For each nominal species, we can then record its taxonomic history — How was it treated by previous authors? This is relevant when trying to incorporate data logged over broad time spans. For example, a museum might contain a record for Lampsilis ovata from Manitoba. Under the current concept of that species, that locality would be far out of range, at other times, the concept of L. ovata also included what is now considered a distinct species, L. cardium, which does occur in the Nelson River drainage of Canada.
Hopefully, it is now apparent how the Genera and Families tables interact as well.
Library Tables
The Publications and Images files are rather simple libraries of bibliographic references and digital images, respectively. Publications records are connected to species, genus and family Descriptions and Syonymies, in the former case to indicate the references where taxon descriptions can be found, and in the latter to indicate the source for a particular taxonomic opinion. Images contains digital photographs of specimens, which are linked to the Specimens file.
In the ERD, the Specimens file is composed of three tables, Specimens, Specimen Identifications and Specimen Images. The need for the latter two tables is simply that there are often multiple IDs and pictures associated with a particular specimen lot; storing the data in a related table allows one-to-many relationships not available in a flat database. In Specimens, we stored catalog numbers, locality, collector, and type data about the shells we handle in the various collections that we have visited. It is through the Specimens file that our specimen images are connected to our concept of actual taxa.
The ERD above is meant to be a schematic of our database. That is, the theory behind it, but in the implementation, we have taken advantage of certain short-cuts and we have also added certain handy tables that are not shown. While the rate of revisions to our system has slowed it has not stopped. In future editions of this web page, when development of the MUSSELpdb is completed, we will be able to provide the full data model and a list of fields. |